Metalloprotein folding and metal site assembly
What is bioinorganic chemistry? Click here
Introduction to my research
My research group explores the factors that control metalloprotein folding and metal site assembly in proteins. Progress in understanding metalloprotein folding has direct significance to human health. Many diseases are characterized or caused by the failure of proteins to fold correctly, and many proteins implicated in folding-related diseases bind redox-active metals. Although group members will not necessarily study proteins involved in human disease, the existence of toxic, misfolded protein conformations illustrates that the folded (native) state of a protein is not necessarily the only biologically relevant conformation. Mutations or abnormal cellular conditions can lead some proteins to populate non-native states.
The characterization of non-native states of proteins in the lab (in vitro) provides valuable insight into conformations that may be accessible to proteins in the cell (in vivo). For example, partially unfolded states we see in the lab may be similar to conformations that are accessible to the protein in the cell and may correspond to trapped states in kinetic folding. Furthermore, the fully unfolded state is the starting point for folding, and identifying structural preferences in the unfolded state may reveal which contacts are important in the early stages of folding. Non-native states of proteins are challenging to characterize, particularly unfolded states, which are "floppy" and are heterogeneous mixtures. Partially unfolded states also may be difficult to characterize; they often make up only a small fraction of the sample, and may be similar to the native or unfolded states. Fortunately for us, many metal-containing proteins are colored, and are therefore great candidates for spectroscopic studies, which can sometimes make structural characterization of non-native states a simpler task than for proteins without metals. In my group, structural characterization of unfolded forms of metalloproteins (non-native states) and refolding from these forms will be used to better understand the process of metalloprotein folding.
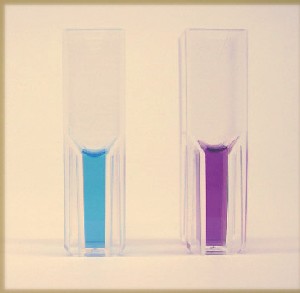
Above: Solutions of two copper-containing proteins: On the left, a "blue copper protein" called azurin, which contains one copper atom. On the right, an engineered azurin variant that binds two copper atoms with a short Cu-Cu distance that gives it an intense purple color. Thanks to Hee Jung Hwang for sharing pictures of her proteins.
Another area of interest in my group is proteins that bind toxic metal ions. See me for more information on this subject.
Research Students
Current students:
- Leigh Clanton (Class of '09)
- Ross Elenkiwich (Class of '10)
- Veronica Taylor (Class of '09)
Past students:
- Meghan Hogdal (Class of '07), chemistry graduate student at Northwestern University
- Pam Nguyen (Class of '07), dental student at AT Still University-Arizona
- Kristin Kaplan (Class of '07), pharmacy student at University of Minnesota-Twin Cities
- Zeb Zacharias (Class of '09)
Joining my group
Researchers in my group will be well served by having completed CHE 107 or a similar course, but beyond that, there are projects available for students of all levels.
Students in my research group will be engaged in some or all of the following activities:
- identification of metal ligands and coordination geometry
- purification of proteins from bacterial cultures
- site-directed mutagenesis
- biochemical and structural characterization of proteins
Techniques we will employ include:
- UV-visible spectroscopy
- 1H, 13C, and 15N NMR spectroscopy
- chromatography
- fluorescence spectroscopy
- gel electrophoresis
- polymerase chain reaction (PCR)
Brandy S. Russell and Kara L. Bren “Denaturant dependence of equilibrium unfolding intermediates and denatured state structure of horse ferricytochrome c.” J. Biol. Inorg. Chem. 2002, 7, 909-916.
Brandy S. Russell, Rory Melenkivitz, and Kara L. Bren. “NMR investigation of ferricytochrome c unfolding: Detection of an unfolding intermediate and residual structure in the denatured state.” Proc. Natl. Acad. Sci., USA 2000, 97, 8312-8317.